Imaging Gut Microbes and Their Surroundings
Observing gut microbes in zebrafish, a model organism, provides insights into how microbial communities nucleate, develop, and interact with each other and with their host. To achieve high-resolution, three-dimensional live imaging in vivo, META researchers have made use of light sheet fluorescence microscopy — [see here], for example. Like all fluorescence-based methods, this technique reports the activities of particular, labeled molecules and species, in our case bacteria engineered to express fluorescent proteins, or fluorescent host cells in transgenic zebrafish. These fluorescent cells cells exist in a landscape of unlabeled tissues and organs that is in general challenging to make sense of.
Though a great deal of effort by research labs around the world has been put into enhancing light sheet fluorescence microscopy, remarkably little work has gone towards combining light sheet imaging with complementary, non-fluorescence-based methods. Differential interference contrast (DIC) microscopy in particular is an ideal target: its utility for imaging transparent, unlabeled specimens such as organisms at early stages of development has been well known for decades.
Spurred by ongoing experiments imaging bacteria in larval zebrafish, META researchers in the Parthasarathy lab realized that an instrument that combines light sheet fluorescence and DIC would be very useful. In DIC imaging, the interference between two slightly displaced optical paths generates contrast, translating gradients in index of refraction into intensity in the final image. This construction can easily be superimposed on the light sheet illumination and detection geometry, simply using different wavelengths for the two modes. The group’s recently published paper [1] describes the setup and its utility. Not only does it allow clear delineation of the boundaries of the larval zebrafish gut, but it allows assessment of the motions of zebrafish cells and of motile bacteria. The gut is a complex environment, and tools that more clearly reveal its structure and behavior will lead to a better understanding of host-microbe interactions.
https://vimeo.com/117772415
*Video Caption:* In this DIC movie, we can see bacteria (here, largely non-motile) in the intestine of a 6 days-post-fertilization larval zebrafish, as well as the gut wall. Examining motile bacteria allows assessment of bacterial speeds, and examining the peristaltic motions of the gut enables characterization of the forces shaping bacterial clusters.
[1] Ryan Baker, Michael J. Taormina, Matthew Jemielita, and Raghuveer Parthasarathy, “A combined light sheet fluorescence and differential interference contrast microscope for live imaging of multicellular specimens,†J. Microscopy [published online ahead of print] (2015). DOI: 10.1111/jmi.12220 [abstract]
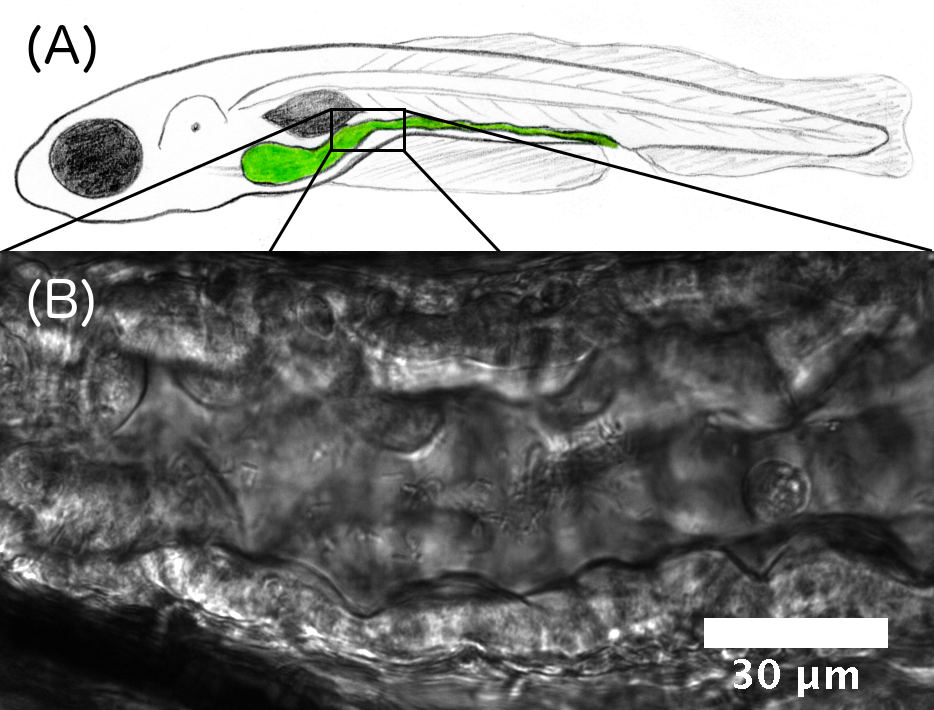
Figure Caption: (A) Schematic illustration of a larval zebrafish, with the intestine highlighted in green. (B) Differential interference contrast image of a section of the intestine. The gut boundaries are clearly evident, as are individual bacteria.
January 27, 2015
