2013 Light Sheet Fluorescence Microscopy Workshop
On Sept. 15-17, META Center PI Raghu Parthasarathy participated in this year’s Light Sheet Fluorescence Microscopy meeting. Light sheet imaging is a central part of the META center’s investigations, as it enables visualization of microbial dynamics inside live zebrafish. As at the past few meetings, talks and discussions spanned both technical advances in light sheet imaging — for example, using electrically tunable lenses to shift focal planes — and applications to a variety of biological systems ranging from zebrafish to flies to cancerous cells to plant roots. Prof. Parthasarathy presented a talk on the META center’s imaging of zebrafish microbial communities, a unique imaging target. This year’s meeting was, for the first time, situated at the U.S. headquarters of Zeiss, which has recently introduced the first commercial light sheet microscope system. It’s a nice, robust microscope that will likely (and hopefully) further increase the impact of light sheet imaging, though, as one would expect, the commercial instrument is limited in its versatility and technical capabilities compared to home-built setups like our own.
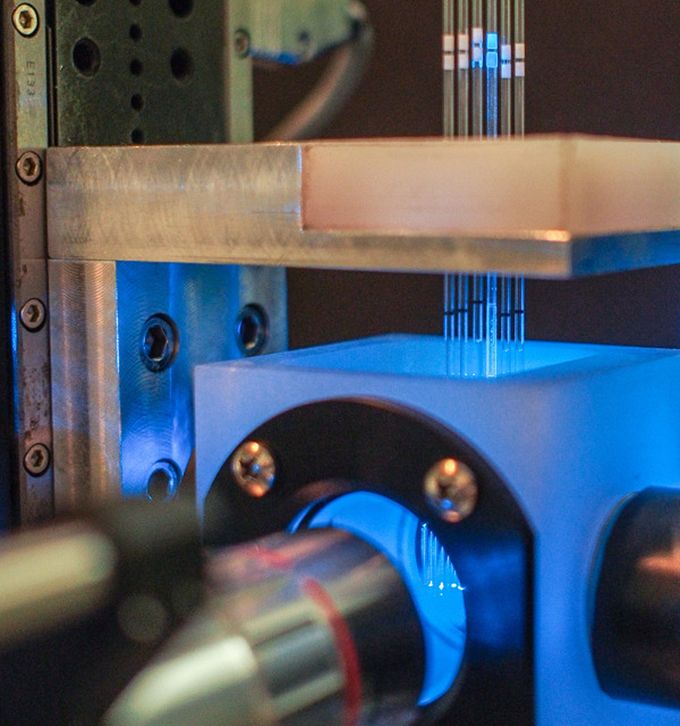
The importance of designing mounting methods that allow imaging of multiple specimens was stressed by Parthasarathy and another group; high-throughput approaches to light sheet imaging will likely be an avenue for future development. Our own specimen holder is pictured; six parallel capillaries each hold a larval fish.
The vast size of light sheet imaging data sets — routinely hundreds of gigabytes or more — and its consequences for computational analysis were the topic of several discussions. New approaches to intelligent image acquisition, real-time processing of data, and other strategies are likely to be a focus of investigation for us, and others, in the future. One discussion touched on whether one should save “raw data” from microscopy experiments (i.e. camera images), or save the output of processing that translates (and condenses) images into some other form. A speaker, arguing for not saving the images themselves, claimed that “raw data” should really mean “the data that gets saved to the hard drive.” Discussion ensued, probably mirroring similar discussions in other areas of science, such as bioinformatics, that are dealing with deluges of data.
October 3, 2013